The Genome Research group employs genomic, molecular biological and bioinformatic methods to elucidate the genetic basis of adaptive variation. We sequence the DNA and RNA of large numbers of individuals to detect sequence variants and differential gene expression. We use these extensive datasets to analyze forest trees' natural genetic variation across the landscape and identify associations with important traits and environmental variables. We are also working on establishing and employing state-of-the-art methods, such as genome editing tools, ultra-long-read sequencing, or pangenomics, in different forest tree species. In conclusion, our work aims to generate novel scientific insights and support political decision making regarding the adaptation of forests under climate change.
Natural genetic variation
Naturally occurring mutations and the resulting genetic variation are essential for driving evolution, domestication and breeding. Natural gene variants provide a great resource for conserving and improving forest ecosystems. For example, alleles conferring field resistance may help to protect tree species challenged by pests and diseases, such as the European ash tree that is severely threatened by an introduced fungus. To this end a thorough understanding of the underlying genetics is crucial. The genetic and molecular elucidation of evolutionary important traits (e.g. dioecy) can furthermore yield insights into fundamental biological processes, and thus inform tree breeding and forest management strategies. Finally, genome-wide adaptive variation can play an important role for forest conservation and adaptation under climate change.
Selected projects:
Poplar dioecy
FraxGen
Beech genomics
Contact: Niels A. Müller
Bioinformatics
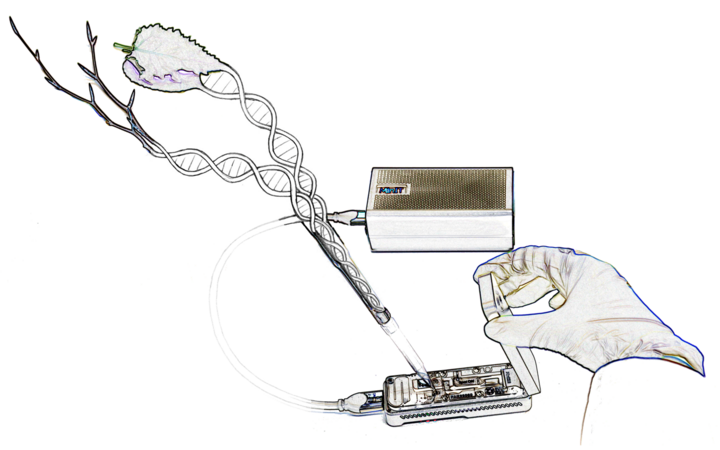
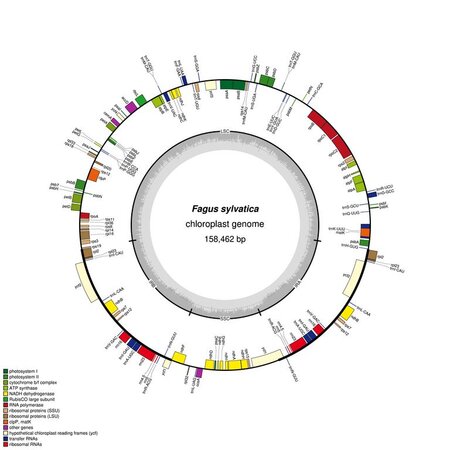
With our bioinformatics team we analyze high-throughput DNA- and RNA-sequencing data in order to develop diagnostic molecular markers and to address functional genomics questions. An important step in many of our analysis pipelines is the genome-wide identification and comparison of genetic variants. For the development of molecular markers for species identification, we also employ whole chloroplast and mitochondrial genomes, some of which we assemble de novo and annotate. Finally we develop databases and bioinformatics tools, to support the institute’s infrastructure.
Selected projects:
TaxGen
Survivor-Oaks
Oak defense
Wood-DNA barcoding
Development of databases
Contact: Birgit Kersten
Genetic Technologies
Genome editing methods, such as CRISPR/Cas, have been significantly developed in crop plants. Further developments are needed for application in different tree species, and these will be used in particular to research the drought stress tolerance of trees.
More information on the group page.
Contact: Tobias Brügmann
Group leader
Scientists
PhD students
Technicians
- Lazic D et al. (2024). Genomic variation of European beech reveals signals of local adaptation despite high levels of phenotypic plasticity. Nature Communications 15, 8553. link
- Leite Montalvão AP et al. (2022) ARR17 controls dioecy in Populus by repressing B-class MADS-box gene expression. Philos Trans Royal Soc B 377 (1850), 20210217. link
- Renner SS, Müller NA (2021) Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nature Plants 7 (4), 392-402. link
- Müller NA et al. (2020) A single gene underlies the dynamic evolution of poplar sex determination. Nature Plants 6 (6), 630-637. link
- Peer-reviewed
- Non peer-reviewed

![[Translate to English:] [Translate to English:]](/media/_processed_/f/3/csm_2022_Titelbild_gross2_Saatgut_in_Hand_9ffb8f5748.jpg)
![[Translate to English:] [Translate to English:]](/media/_processed_/f/3/csm_2022_Titelbild_gross2_Saatgut_in_Hand_c17270fcc0.jpg)